NCBI provides an easy way to search DNA/Amino acid sequence databases that DO NOT match to a target organisms genome, yet match to other databases and sources.
Just use the exclude field in NCBI's BLASTn
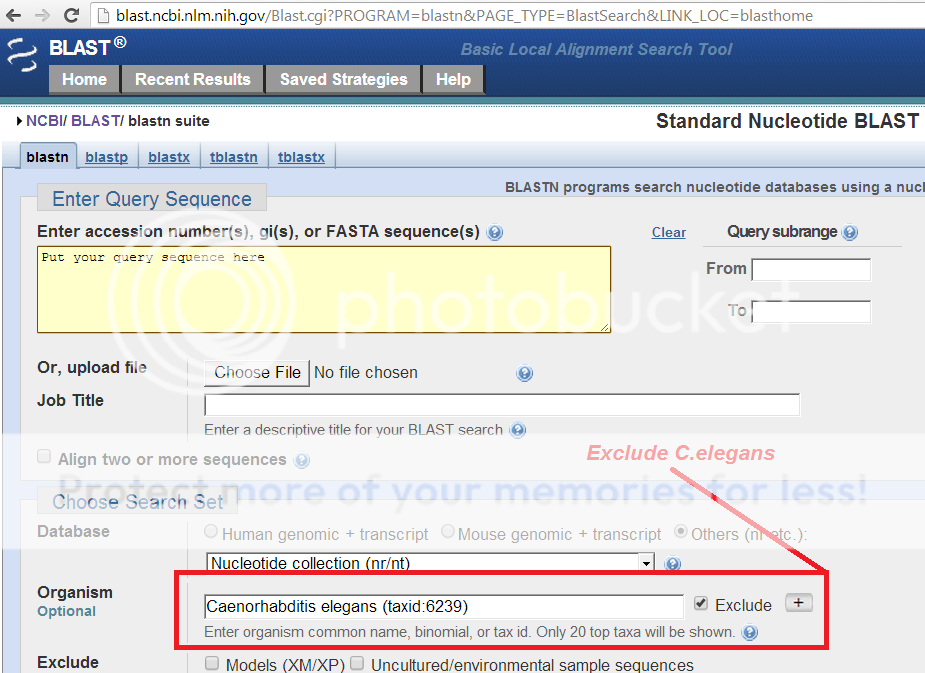
Hope this was useful.
Solución
NCBI provides an easy way to search DNA/Amino acid sequence databases that DO NOT match to a target organisms genome, yet match to other databases and sources.
Just use the exclude field in NCBI's BLASTn

Hope this was useful.